The transcriptional profiles of these individual cells coupled with assembled t cell receptor tcr sequences enable us to identify 11 t cell subsets based on.
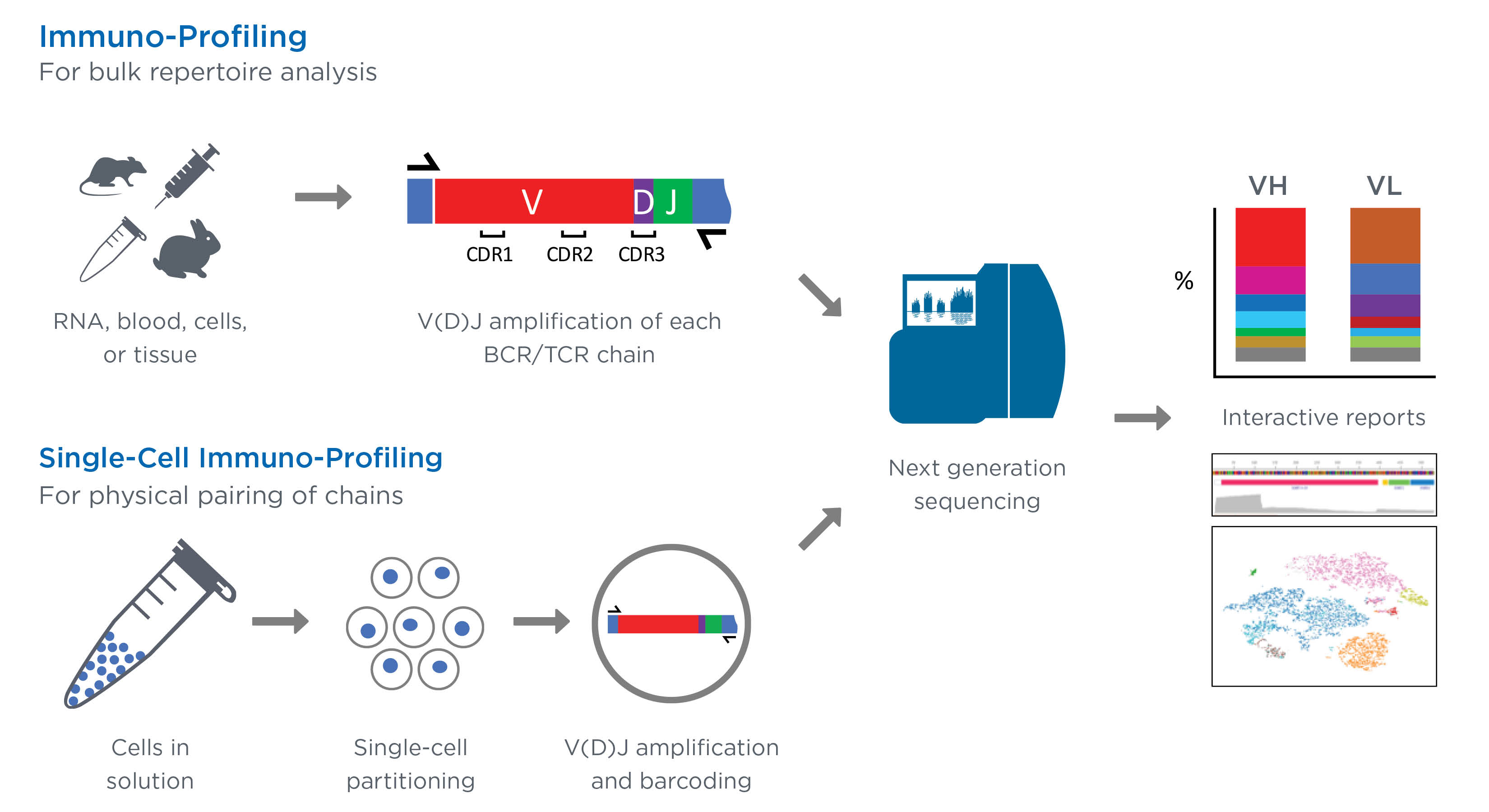
Single cell tcr sequencing.
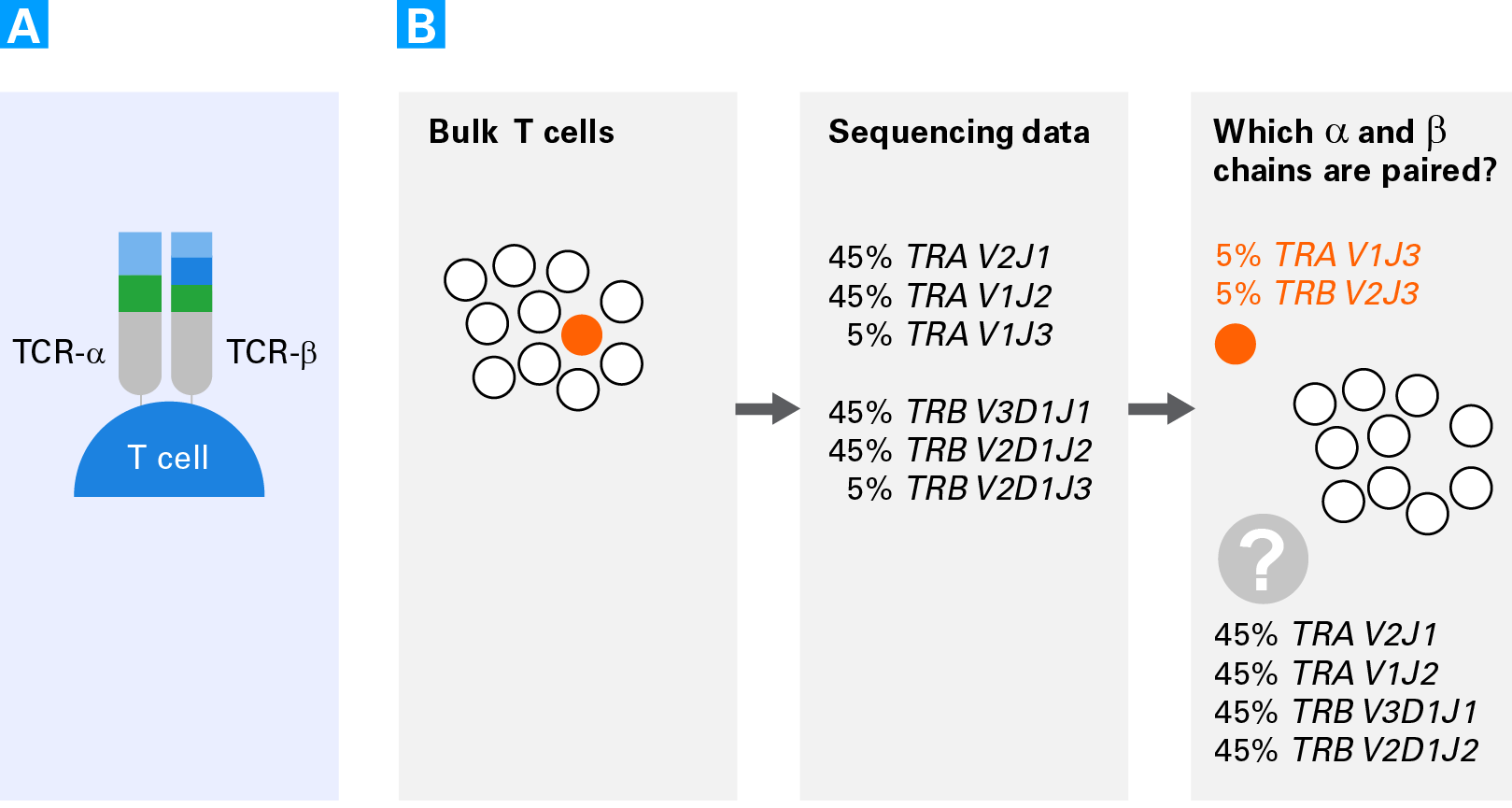
However current methods for tcr inference from scrna seq are limited in their sensitivity and require long sequencing reads thus increasing the cost and decreasing the number of cells that can be.
Single cell v d j cell surface protein v1 and v1 1 minimum sequencing depth.
The recent development of single cell rna sequencing scrnaseq allows the transcriptomes of thousands of cells to be processed simultaneously bringing a way to identify subpopulations of cells and provide functional insights such as the identification of each cell s unique tcrs and paired alpha and beta heterodimers that were previously.
We do not recommend sequencing these libraries with a dual index configuration.
Clonal replacement of tumor specific t cells following pd 1 blockade.
Single cell sequencing examines the sequence information from individual cells with optimized next generation sequencing ngs technologies providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment.
Here we perform deep single cell rna sequencing on 5 063 single t cells isolated from peripheral blood tumor and adjacent normal tissues from six hepatocellular carcinoma patients.
T cells are the core components of our adaptive immune system.
Once activated they can directly kill cells that are foreign cytolytic t cells or perform helper function helper t cells to activate b cells to make antibodies against foreign antigen.
Our results support a model of continuous activation in t cells and do not comport with the macrophage polarization model in cancer.
Single cell v d j cell surface protein v1 and v1 1 libraries are single indexed.
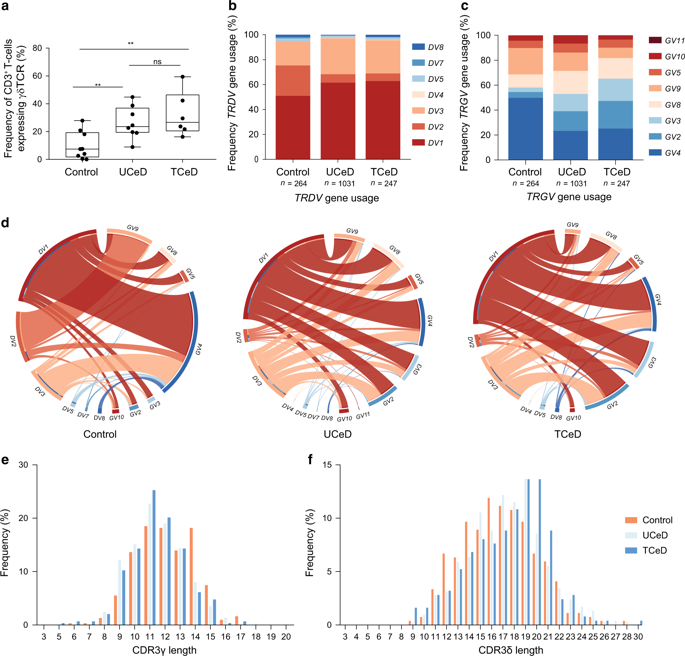
Analysis of paired single cell rna and t cell receptor tcr sequencing data from 27 000 additional t cells revealed the combinatorial impact of tcr utilization on phenotypic diversity.
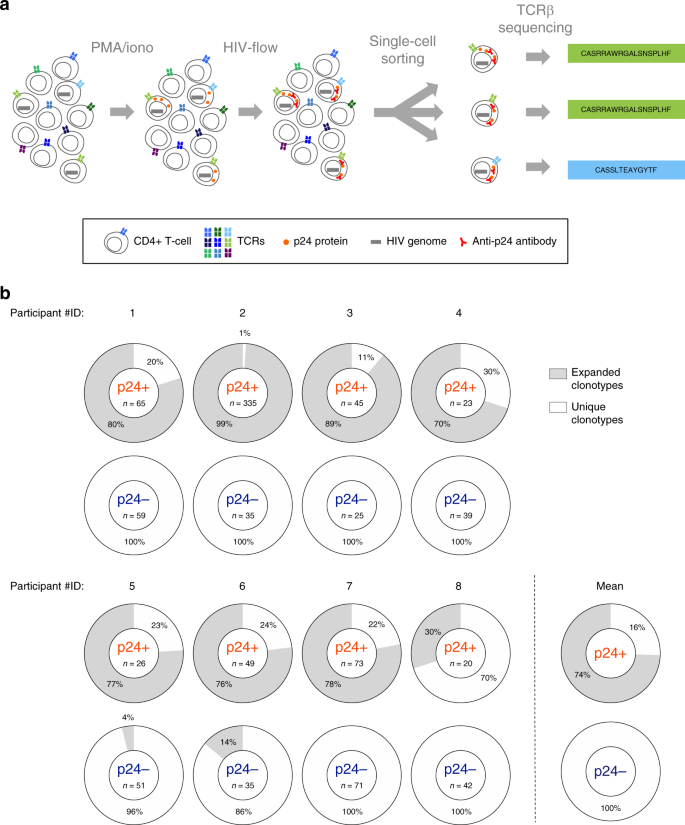
To assess the expressed tcr repertoire of phenotypically defined mouse t cell populations at single cell level we designed a strategy for the high throughput amplification and sequencing of paired tcrα and tcrβ genes.
Single cell based approaches brought the analysis to a higher level of complexity and now provide the opportunity to sequence paired alpha and beta chains.
We also discuss novel approaches that through the integration of tcr tracking and mrna single cell sequencing offer a valuable tool to associate antigen specificity to transcriptional.
For example in cancer sequencing the dna of individual cells can give information about.